
gget-mcp
Bioinformatics tools via Model Context Protocol for AI-powered genomics analysis.
Key Features
Use Cases
README
gget-mcp
MCP (Model Context Protocol) server for the gget bioinformatics library.
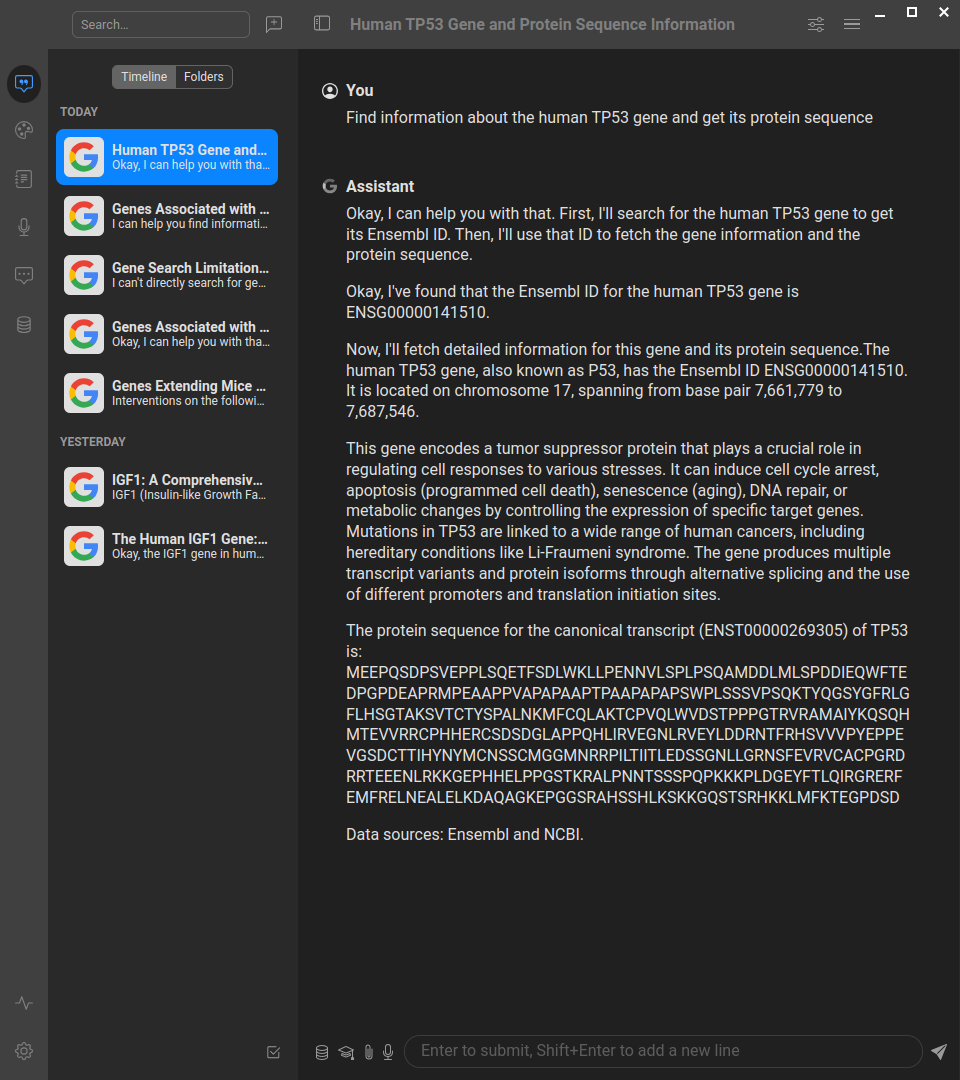
This server implements the Model Context Protocol (MCP) for gget, providing a standardized interface for accessing powerful bioinformatics tools and databases. MCP enables AI assistants and agents to perform complex genomics queries through structured interfaces.
The gget bioinformatics toolkit provides efficient querying of genomic databases and includes functions for:
- Gene and sequence information retrieval
- BLAST and sequence alignment
- Protein structure prediction
- Gene expression analysis
- Functional enrichment analysis
- And much more!
🚀 Quick Example: What Can You Do?
👤 User: "Analyze the TP53 gene - find its details and protein sequence, then check for cancer mutations"
🤖 AI Assistant: I'll help you analyze TP53 step by step:
[Uses gget_search] → Found TP53 (ENSG00000141510) on chromosome 17
[Uses gget_info] → TP53 is a protein-coding tumor suppressor gene
[Uses gget_seq with translate=True] → Retrieved 393 amino acid protein sequence
[Uses gget_cosmic] → Found hotspot mutations R175H, R248Q, R273H in multiple cancers
👤 User: "Now predict the 3D structure of this protein sequence: MSKGEELFTGVVPILVEL..."
🤖 AI Assistant: Let me analyze this sequence:
[Uses gget_blast] → Identified as Green Fluorescent Protein (GFP)
[Uses gget_alphafold] → Predicted classic beta-barrel structure with 90%+ confidence
👤 User: "Perform pathway enrichment for DNA repair genes: TP53, BRCA1, BRCA2, ATM"
🤖 AI Assistant: [Uses gget_enrichr] → Top pathways: DNA Damage Response, Homologous Recombination, Cell Cycle Checkpoints, p53 Signaling
See how powerful this is? You can now ask natural language questions about genes, sequences, mutations, and pathways - the AI does the complex bioinformatics work for you!
About MCP (Model Context Protocol)
MCP is a protocol that bridges the gap between AI systems and specialized domain tools. It enables:
- Structured Access: Direct connection to powerful bioinformatics tools and databases
- Natural Language Queries: Simplified interaction with complex genomics workflows
- Type Safety: Strong typing and validation through FastMCP
- AI Integration: Seamless integration with AI assistants and agents
If you want to understand more about what the Model Context Protocol is and how to use it more efficiently, you can take the DeepLearning AI Course or search for MCP videos on YouTube.
Available Tools
Gene Information & Search
gget_search: Find Ensembl IDs associated with search termsgget_info: Fetch detailed information for Ensembl IDsgget_seq: Retrieve nucleotide or amino acid sequencesgget_ref: Get reference genome information from Ensembl
Sequence Analysis
gget_blast: BLAST nucleotide or amino acid sequencesgget_blat: Find genomic locations of sequencesgget_muscle: Align multiple sequences
Expression & Functional Analysis
gget_archs4: Get gene expression data from ARCHS4gget_enrichr: Perform gene set enrichment analysis
Protein Structure & Function
gget_pdb: Fetch protein structure data from PDBgget_alphafold: Predict protein structure using AlphaFold
Cancer & Mutation Analysis
gget_cosmic: Search COSMIC database for cancer mutations
Single-cell Analysis
gget_cellxgene: Query single-cell RNA-seq data from CellxGene
Quick Start
# Download and install uv
curl -LsSf https://astral.sh/uv/install.sh | sh
# Verify installation
uv --version
uvx --version
uvx is a very nice tool that can run a python package installing it if needed.
Running with uvx
You can run the gget-mcp server directly using uvx without cloning the repository:
# Run the server in HTTP mode (default)
uvx gget-mcp http
STDIO Mode (for MCP clients that require stdio)
# Run the server in stdio mode
uvx gget-mcp stdio
HTTP Mode (Web Server)
# Run the server in streamable HTTP mode on default (3002) port
uvx gget-mcp http
# Run on a specific port
uvx gget-mcp http --port 8000
SSE Mode (Server-Sent Events)
# Run the server in SSE mode
uvx gget-mcp sse
In cases when there are problems with uvx often they can be caused by cleaning uv cache:
uv cache clean
The HTTP mode will start a web server that you can access at http://localhost:3002/mcp (with documentation at http://localhost:3002/docs). The STDIO mode is designed for MCP clients that communicate via standard input/output, while SSE mode uses Server-Sent Events for real-time communication.
Note: Currently, we do not have a Swagger/OpenAPI interface, so accessing the server directly in your browser will not show much useful information. To explore the available tools and capabilities, you should either use the MCP Inspector (see below) or connect through an MCP client to see the available tools.
Configuring your AI Client (Anthropic Claude Desktop, Cursor, Windsurf, etc.)
We provide preconfigured JSON files for different use cases. Here are the actual configuration examples:
STDIO Mode Configuration (Recommended)
Use this configuration for most AI clients. Use this mode when you want to save large output files (sequences, structures, alignments) to disk instead of returning them as text. Create or update your MCP configuration file:
{
"mcpServers": {
"gget-mcp": {
"command": "uvx",
"args": ["--from", "gget-mcp@latest", "stdio"]
}
}
}
HTTP Mode Configuration
For HTTP mode:
{
"mcpServers": {
"gget-mcp": {
"command": "uvx",
"args": ["--from", "gget-mcp@latest", "server"]
}
}
}
Configuration Video Tutorial
For a visual guide on how to configure MCP servers with AI clients, check out our configuration tutorial video for our sister MCP server (biothings-mcp). The configuration principles are exactly the same for the gget MCP server - just use the appropriate JSON configuration files provided above.
Inspecting gget MCP server
If you want to inspect the methods provided by the MCP server, use npx (you may need to install nodejs and npm):
For STDIO mode with uvx:
npx @modelcontextprotocol/inspector --config mcp-config.json --server gget-mcp
You can also run the inspector manually and configure it through the interface:
npx @modelcontextprotocol/inspector
After that you can explore the tools and resources with MCP Inspector at which is usually at 6274 port (note, if you run inspector several times it can change port)
Integration with AI Systems
Simply point your AI client (like Cursor, Windsurf, ClaudeDesktop, VS Code with Copilot, or others) to use the appropriate configuration file from the repository.
Repository setup
# Clone the repository
git clone https://github.com/longevity-genie/gget-mcp.git
cd gget-mcp
uv sync
Running the MCP Server
If you already cloned the repo you can run the server with uv:
# Start the MCP server locally (HTTP mode)
uv run server
# Or start in STDIO mode
uv run stdio
# Or start in SSE mode
uv run sse
Safety Features
- Input validation: Comprehensive parameter validation for all gget functions
- Error handling: Robust error handling with informative messages
- Rate limiting: Respectful usage of external APIs and databases
- Data validation: Type checking and data format validation
Testing & Verification
The MCP server is provided with comprehensive tests including both unit tests and integration tests for network-dependent operations.
Running Tests
Run tests for the MCP server:
# Run all tests (excluding expensive judge tests)
uv run pytest -vvv -m "not judge"
# Run only fast tests (skip slow, integration, and judge tests)
uv run pytest -vvv -m "not slow and not integration and not judge"
# Run only integration tests
uv run pytest -vvv -m integration
# Run judge tests (expensive LLM tests - requires API keys and may cost money)
uv run pytest test/test_judge.py -vvv
# Run tests with coverage (excluding judge tests)
uv run pytest -vvv --cov=src/gget_mcp --cov-report=term-missing -m "not judge"
Note on Judge Tests: The judge tests (test_judge.py) use large language models to evaluate the AI agent's performance with gget tools. These tests:
- Are automatically excluded from CI/CD to avoid costs
- Require API keys (GEMINI_API_KEY or similar)
- May incur charges from LLM API providers
- Are designed for local development and manual evaluation
- Provide valuable insights into how well the tools work with AI agents
GitHub Actions Workflows
This project includes several GitHub Actions workflows:
- CI (
.github/workflows/ci.yml): Runs on every push/PR with basic linting and fast tests - Tests (
.github/workflows/test.yml): Comprehensive testing across multiple OS and Python versions- Fast tests on all platforms
- Integration tests (may require network access)
- Slow tests (like BLAST operations) - runs only on Ubuntu with Python 3.11
- Code coverage reporting
- Security checks with bandit and safety
- Publish (
.github/workflows/publish.yml): Publishes to PyPI when tags are pushed
You can use MCP inspector with locally built MCP server same way as with uvx.
Note: Using the MCP Inspector is optional. Most MCP clients (like Cursor, Windsurf, etc.) will automatically display the available tools from this server once configured. However, the Inspector can be useful for detailed testing and exploration.
If you choose to use the Inspector via npx, ensure you have Node.js and npm installed. Using nvm (Node Version Manager) is recommended for managing Node.js versions.
Example Questions from Test Suite
Here are validated example questions that you can ask the AI assistant when using this MCP server:
- "Find information about the human TP53 gene and get its protein sequence."
- "What are the orthologs of BRCA1 gene across different species?"
- "Perform enrichment analysis for a set of cancer-related genes: TP53, BRCA1, BRCA2, ATM, CHEK2."
- "Get the 3D structure information for the protein encoded by the EGFR gene."
- "Find mutations in the COSMIC database for the PIK3CA gene."
- "Analyze gene expression patterns for insulin (INS) gene across different tissues."
- "Perform BLAST search with a DNA sequence to identify its origin: ATGGCGCCCGAACAGGGAC."
- "Find diseases associated with the APOE gene using OpenTargets."
- "Get reference genome information for mouse (Mus musculus)."
- "Align multiple protein sequences and identify conserved regions."
These questions are taken directly from our automated test suite and are validated to work correctly with the available gget tools.
Contributing
We welcome contributions from the community! 🎉 Whether you're a researcher, developer, or enthusiast interested in bioinformatics and genomics research, there are many ways to get involved:
We especially encourage you to try our MCP server and share your feedback with us! Your experience using the server, any issues you encounter, and suggestions for improvement are incredibly valuable for making this tool better for the entire research community.
Ways to Contribute
- 🐛 Bug Reports: Found an issue? Please open a GitHub issue with detailed information
- 💡 Feature Requests: Have ideas for new functionality? We'd love to hear them!
- 📝 Documentation: Help improve our documentation, examples, or tutorials
- 🧪 Testing: Add test cases, especially for edge cases or new query patterns
- 🔍 Data Quality: Help identify and report data inconsistencies or suggest improvements
- 🚀 Performance: Optimize queries, improve caching, or enhance server performance
- 🌐 Integration: Create examples for new MCP clients or AI systems
- 🎥 Tutorials & Videos: Create tutorials, video guides, or educational content showing how to use MCP servers
- 📖 User Stories: Share your research workflows and success stories using our MCP servers
- 🤝 Community Outreach: Help us evangelize MCP adoption in the bioinformatics community
Tutorials, videos, and user stories are especially valuable to us! We're working to push the bioinformatics community toward AI adoption, and real-world examples of how researchers use our MCP servers help demonstrate the practical benefits and encourage wider adoption.
Getting Started
- Fork the repository
- Create a feature branch (
git checkout -b feature/amazing-feature) - Make your changes and add tests
- Run the test suite (
uv run pytest) - Commit your changes (
git commit -m 'Add amazing feature') - Push to your branch (
git push origin feature/amazing-feature) - Open a Pull Request
Development Guidelines
- Follow the existing code style (we use
rufffor formatting and linting) - Add tests for new functionality
- Update documentation as needed
- Keep commits focused and write clear commit messages
Questions or Ideas?
Don't hesitate to open an issue for discussion! We're friendly and always happy to help newcomers get started. Your contributions help advance open science and bioinformatics research for everyone. 🧬✨
Known Issues
External Dependencies
Some gget functions depend on external web services and databases that may occasionally be unavailable or rate-limited. The server implements proper error handling and retries where appropriate.
Test Coverage
While we provide comprehensive tests including integration tests for network-dependent operations, some test cases may be sensitive to external service availability. Some automated test results may need manual validation.
License
This project is licensed under the MIT License.
Acknowledgments
- gget - The amazing bioinformatics toolkit this server wraps
- FastMCP - The MCP server framework used
- Model Context Protocol - The protocol specification
Other MCP Servers by Longevity Genie
We also develop other specialized MCP servers for biomedical research:
- biothings-mcp - MCP server for BioThings.io APIs, providing access to gene annotation (mygene.info), variant annotation (myvariant.info), and chemical compound data (mychem.info)
- opengenes-mcp - MCP server for OpenGenes database, providing access to aging and longevity research data
- synergy-age-mcp - MCP server for SynergyAge database, providing access to synergistic and antagonistic longevity gene interactions
We are supported by:
HEALES - Healthy Life Extension Society
and
IBIMA - Institute for Biostatistics and Informatics in Medicine and Ageing Research
Star History
Repository Owner
Organization
Repository Details
Programming Languages
Tags
Join Our Newsletter
Stay updated with the latest AI tools, news, and offers by subscribing to our weekly newsletter.
Related MCPs
Discover similar Model Context Protocol servers

awslabs/mcp
Specialized MCP servers for seamless AWS integration in AI and development environments.
AWS MCP Servers is a suite of specialized servers implementing the open Model Context Protocol (MCP) to bridge large language model (LLM) applications with AWS services, tools, and data sources. It provides a standardized way for AI assistants, IDEs, and developer tools to access up-to-date AWS documentation, perform cloud operations, and automate workflows with context-aware intelligence. Featuring a broad catalog of domain-specific servers, quick installation for popular platforms, and both local and remote deployment options, it enhances cloud-native development, infrastructure management, and workflow automation for AI-driven tools. The project includes Docker, Lambda, and direct integration instructions for environments such as Amazon Q CLI, Cursor, Windsurf, Kiro, and VS Code.
- ⭐ 6,220
- MCP
- awslabs/mcp

mcp
Universal remote MCP server connecting AI clients to productivity tools.
WayStation MCP acts as a remote Model Context Protocol (MCP) server, enabling seamless integration between AI clients like Claude or Cursor and a wide range of productivity applications, such as Notion, Monday, Airtable, Jira, and more. It supports multiple secure connection transports and offers both general and user-specific preauthenticated endpoints. The platform emphasizes ease of integration, OAuth2-based authentication, and broad app compatibility. Users can manage their integrations through a user dashboard, simplifying complex workflow automations for AI-powered productivity.
- ⭐ 27
- MCP
- waystation-ai/mcp

mcpmcp-server
Seamlessly discover, set up, and integrate MCP servers with AI clients.
mcpmcp-server enables users to discover, configure, and connect MCP servers with preferred clients, optimizing AI integration into daily workflows. It supports streamlined setup via JSON configuration, ensuring compatibility with various platforms such as Claude Desktop on macOS. The project simplifies the connection process between AI clients and remote Model Context Protocol servers. Users are directed to an associated homepage for further platform-specific guidance.
- ⭐ 17
- MCP
- glenngillen/mcpmcp-server

mcp-server-templates
Deploy Model Context Protocol servers instantly with zero configuration.
MCP Server Templates enables rapid, zero-configuration deployment of production-ready Model Context Protocol (MCP) servers using Docker containers and a comprehensive CLI tool. It provides a library of ready-made templates for common integrations—including filesystems, GitHub, GitLab, and Zendesk—and features intelligent caching, smart tool discovery, and flexible configuration options via JSON, YAML, environment variables, or CLI. Perfect for AI developers, data scientists, and DevOps teams, it streamlines the process of setting up and managing MCP servers and has evolved into the MCP Platform for enhanced capabilities.
- ⭐ 5
- MCP
- Data-Everything/mcp-server-templates

OpenMCP
A standard and registry for converting web APIs into MCP servers.
OpenMCP defines a standard for converting various web APIs into servers compatible with the Model Context Protocol (MCP), enabling efficient, token-aware communication with client LLMs. It also provides an open-source registry of compliant servers, allowing clients to access a wide array of external services. The platform supports integration with local and remote hosting environments and offers tools for configuring supported clients, such as Claude desktop and Cursor. Comprehensive guidance is offered for adapting different API formats including REST, gRPC, GraphQL, and more into MCP endpoints.
- ⭐ 252
- MCP
- wegotdocs/open-mcp
Didn't find tool you were looking for?




